Multi contrast, 2D and 3D
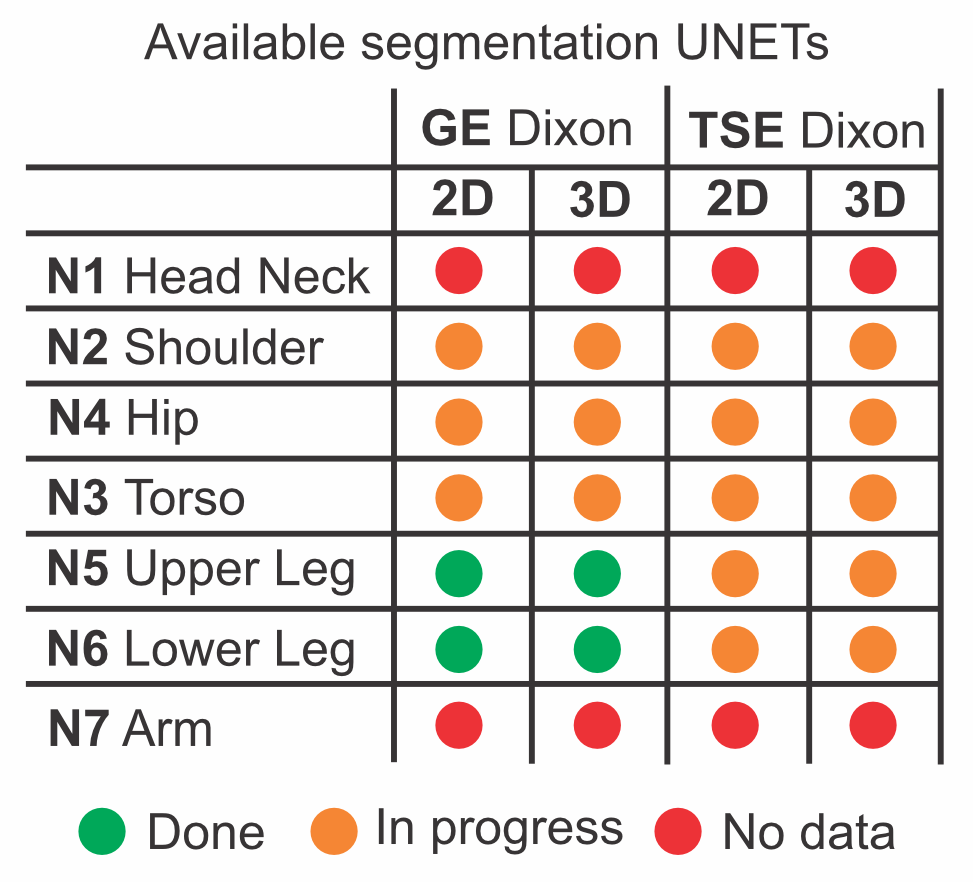
The available segmentation networks
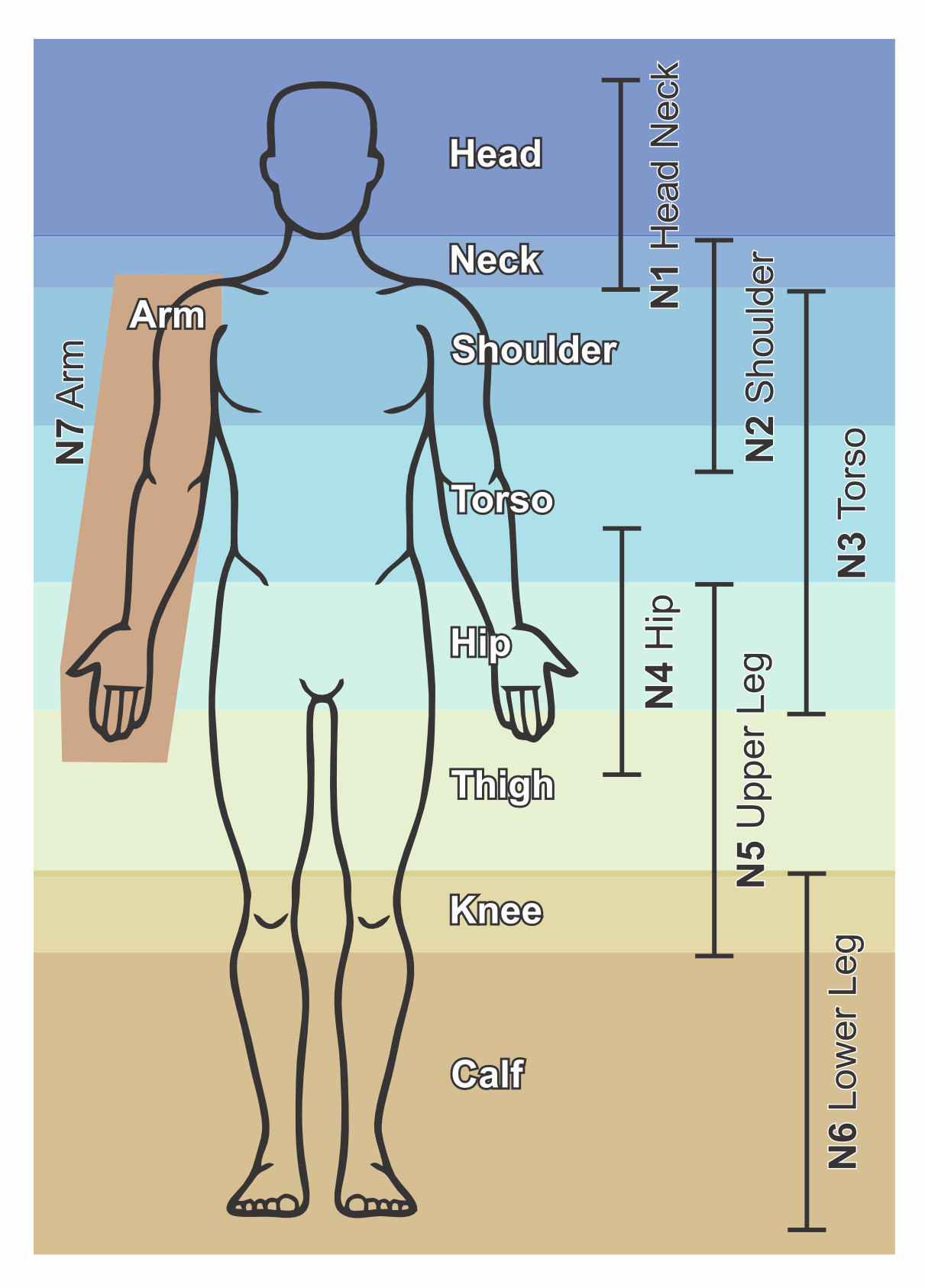
With our framework we are able to automatically detect the body location. For each body part dedicated small light weight networks are developed and trained. If the data contains multiple locations the data is cut in various regions with considerable overlap and each is segmented by the correct neural network after which everything is collected in one segmentations file.
We aim to have segmentations networks for both GE and TSE Dixon based imaging that work on the raw magnitude data, in and out phase images and the reconstructed water and fat maps. Segmentation works best on 3D continuous datasets however those are not always available. Therefore we train all our networks in 2D and 3D. If data is acquired in 2D with slice gaps or extremely thick slices the 2D networks can be used. However 3D continuous data with 3D segmentation networks outperform the 2D variants consistently.
- How to use the muscle segmentation framwork
- Muscle segmentation naming
- The available segmentation networks
- Script based segmentation without mathematica
- Automated muscle segmentation
- Tools for muscle segmentation
We gratefully acknowledge the support of NVIDIA with the donation of GPU used for this work.