Why QMRITools uses Muscle-BIDS
Muscle-BIDS
Why Muscle-BIDS
For automated data processing of processing with various tool-boxes data management is important. For this, the muscle MRI community is developing the Muscle BIDS data structure. When data is organized in this way all needed parameters and data sets can automatically be found, which enables automated processing and evaluation of large data sets. Furthermore, each toolbox that is Muscle-BIDS compatible can run its analysis on the same data without the need to restructure the data input. The last advantage is that the output is also standardized which makes the comparison of results and methods more convenient. More about Muscle-Bids can be found here».
QMRITools and Muscle-BIDS
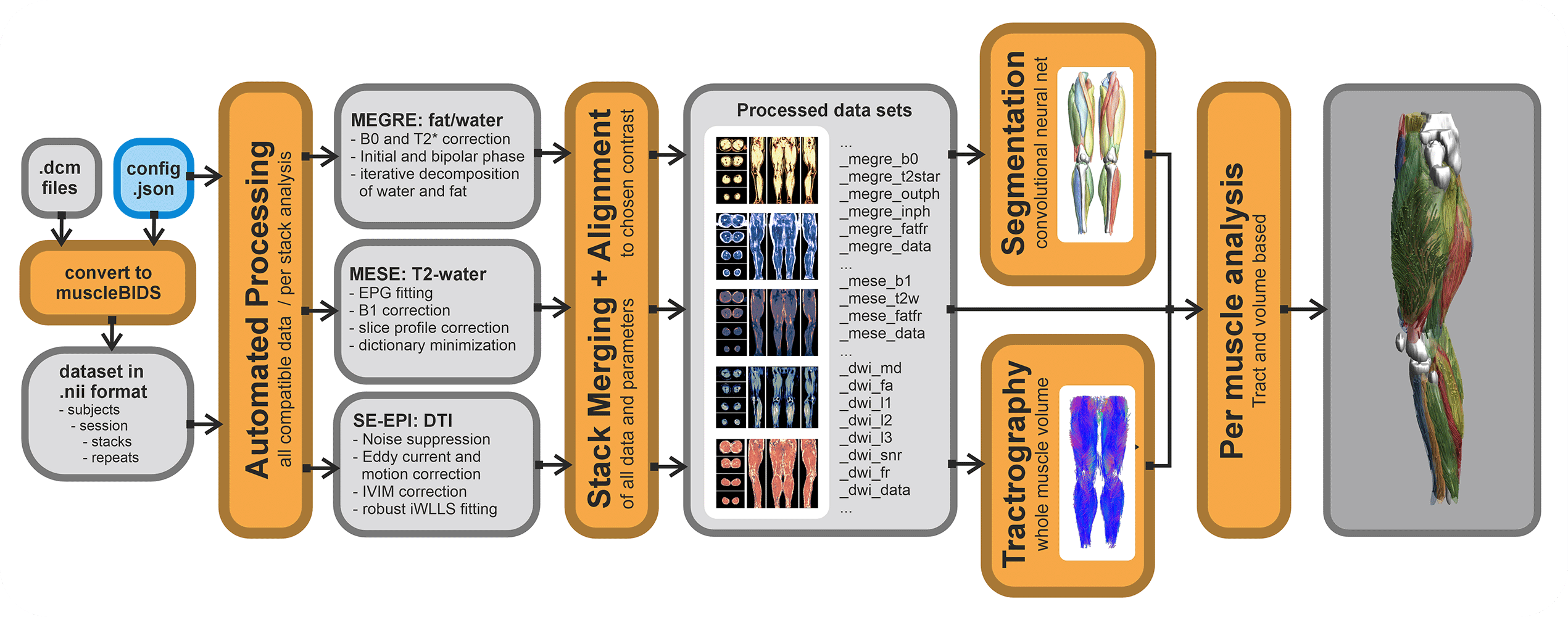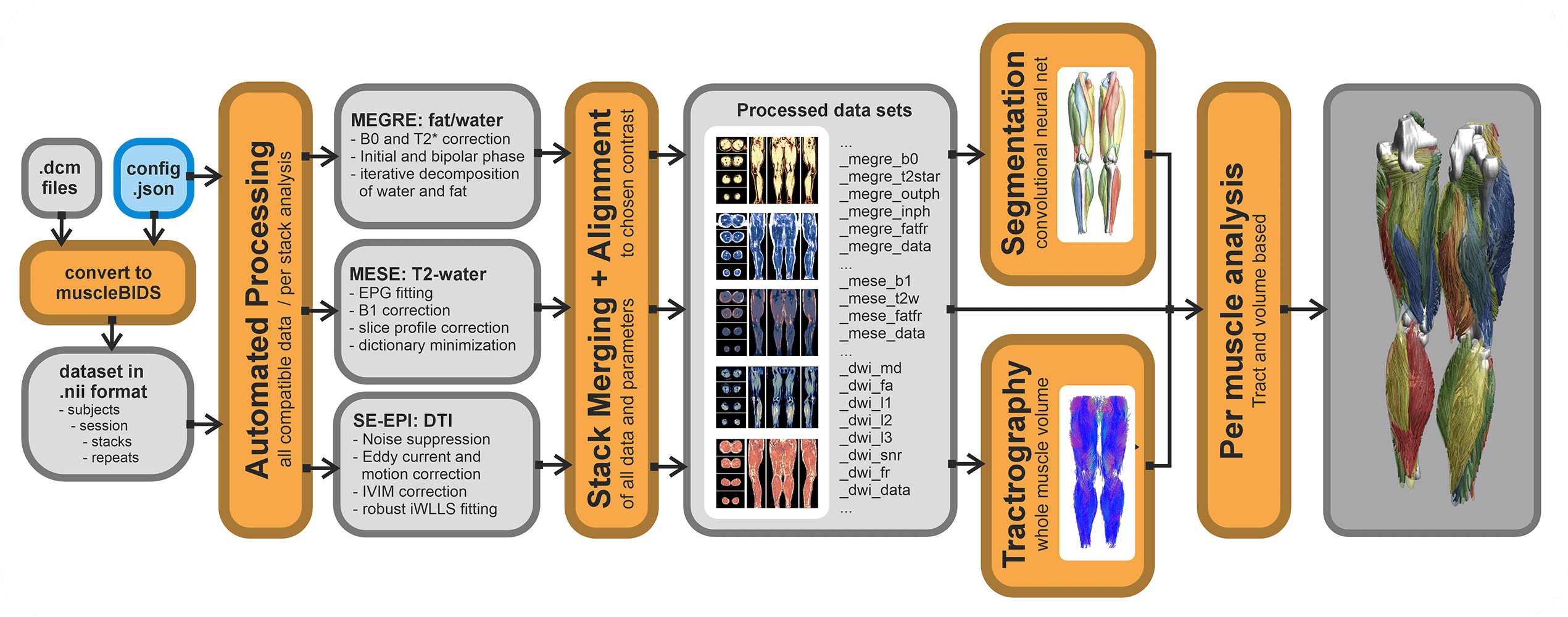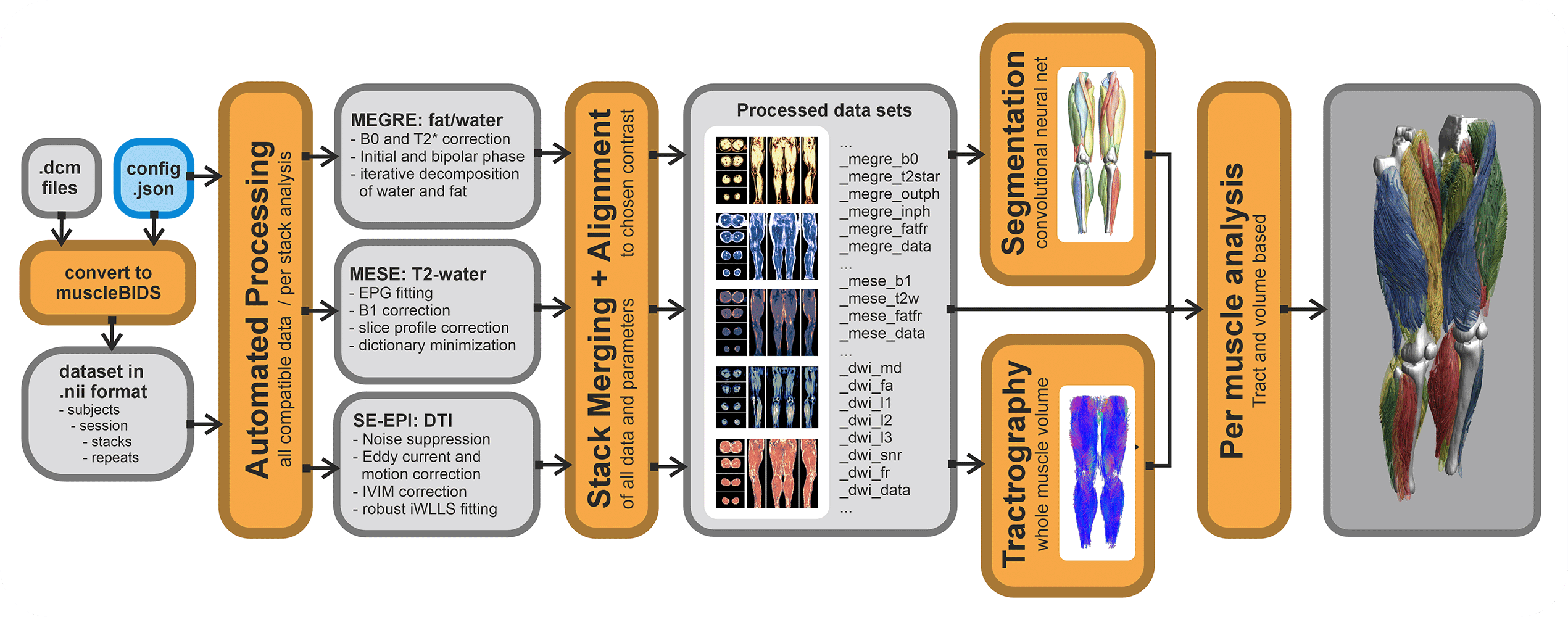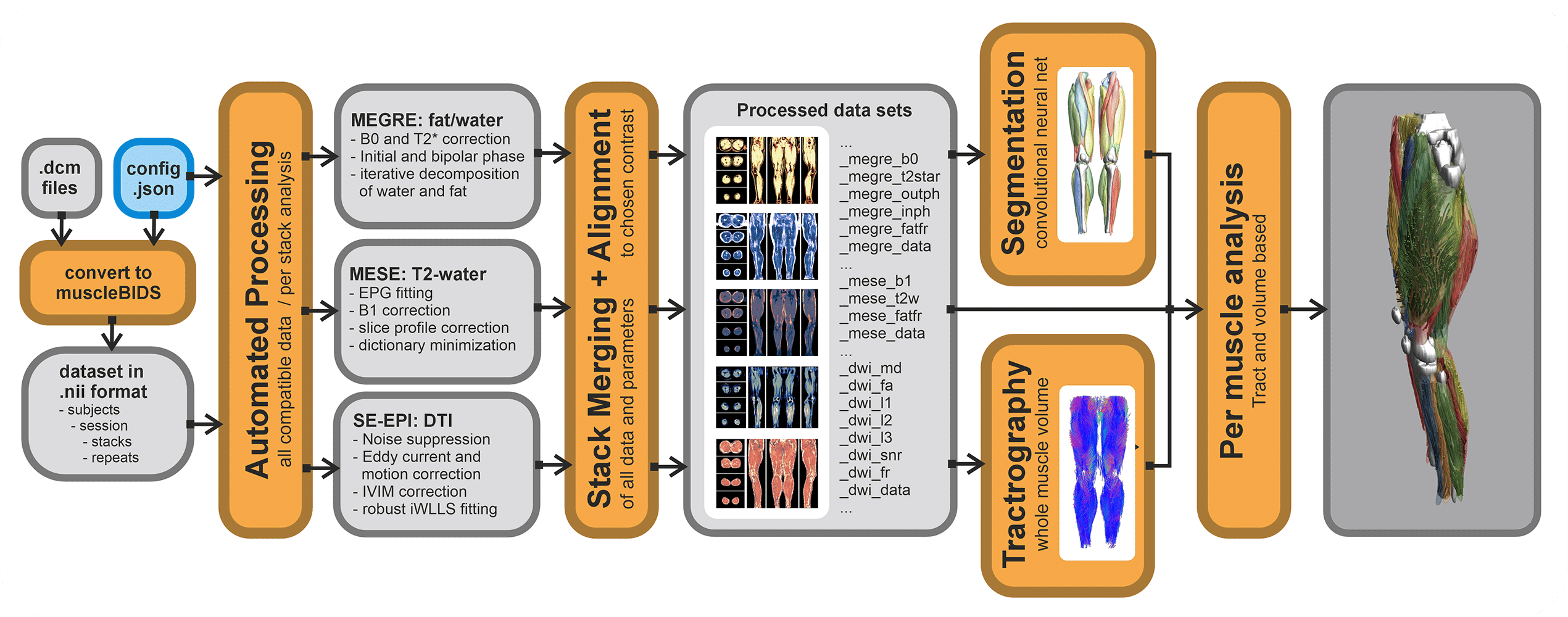
QMRITools contains a couple of Muscle-BIDS scripts. Of course, all functionality of QMRITools remains available without the use of Muscle-BIDS but for standardized qMRI muscle studies using the Muscle-BIDS conventions can save lots of time and fully automated scripts are available for common data processing methods. Muscle-BIDS pipelines can support the following steps:
- Conversion of DICOM to raw Muscle-BIDS NIfTI
- Processing of Muscle-BIDS data
- Analysis of processed Muscle-BIDS data
- Conversion of Muscle-BIDS output to DICOM
Currently, QMRITools supports step 1 for Philips MRI data if the data acquisition is performed in a standardized way. Furthermore, it fully supports steps 2 and 3 for DIXON, T2 and DTI processing and analysis. On this page, for each step, it is explained how QMRITools handles Muscle-BIDS data and what function can be used. Eventually, the aim is to be able to fully automate steps 1 to 4 using stand-alone scripts that only need to be configured once per study or data set.
General guidelines
Within QMRITools a Muscle-BIDS dataset is composed of:
- One NIfTI file containing the image data.
- One JSON header named identical to the NIfTI file containing the imaging parameters relevant to the specific acquisition type and processing.
Multi-slice 2D acquisitions are saved as a 3D NIfTI volume. Where the voxel size stored in the Nifti file is the slice spacing rather than the actual slice thickness. In the header, the actual slice thickness and slice spacing are stored. To reduce the number of files, where appropriate, 4D NIfTI datasets are used instead of multiple 3D files (for example, multi-echo or diffusion data). The quantity that changes in the fourth dimension is identified by the parameter FourthDimension in the header. E.g. "FourthDimension": "EchoTime". The value of the FourthDimension field must have a corresponding entry in the header, which is a list of values corresponding to each index along the fourth dimension. E.g. "EchoTime": [3.9, 6.3].
A Muscle-BIDS dataset must contain the source data, which in this case is the unprocessed nifty data with its headers. Each unique subject has its folder. Within the subject folder, there are session folders for when the same subject is scanned on multiple occasions. Within the session folder, each datatype is again stored in its subfolders.
QMRITools add various other logging and file tree files to each folder that are not mandatory but can be useful and are sometimes needed for processing. An example of a Muscle-BIDS folder structure is given below:
├─sub-Test001/
│ └─ses-001/
│ ├─dix/
│ │ ├─sub-Test001_ses-001_stk-DIXON1_megre_imag.json
│ │ ├─sub-Test001_ses-001_stk-DIXON1_megre_imag.nii.gz
│ │ ├─sub-Test001_ses-001_stk-DIXON1_megre.json
│ │ ├─..<<34>>..
│ │ ├─sub-Test001_ses-001_stk-DIXON5_megre_ph.nii.gz
│ │ ├─sub-Test001_ses-001_stk-DIXON5_megre_real.json
│ │ └─sub-Test001_ses-001_stk-DIXON5_megre_real.nii.gz
│ ├─dwi/
│ │ ├─sub-Test001_ses-001_stk-DTI1_dwi_dti.bval
│ │ ├─sub-Test001_ses-001_stk-DTI1_dwi_dti.bvec
│ │ ├─sub-Test001_ses-001_stk-DTI1_dwi_dti.json
│ │ ├─..<<14>>..
│ │ ├─sub-Test001_ses-001_stk-DTI5_dwi_dti.bvec
│ │ ├─sub-Test001_ses-001_stk-DTI5_dwi_dti.json
│ │ └─sub-Test001_ses-001_stk-DTI5_dwi_dti.nii.gz
│ ├─raw/
│ │ ├─101_20221213162506_20221212-set1_Ph_20221212-set1_SURVEY_2_LEG_GR.json
│ │ ├─101_20221213162506_20221212-set1_Ph_20221212-set1_SURVEY_2_LEG_GR.nii.gz
│ │ ├─201_20221213162506_20221212-set1_Ph_20221212-set1_SURVEY_2_LEG_GR.json
│ │ ├─..<<512>>..
│ │ ├─905_20221213162506_20221212-set1_Ph_20221212-set1_WIP_DTI5_SE.json
│ │ ├─905_20221213162506_20221212-set1_Ph_20221212-set1_WIP_DTI5_SE.nii.gz
│ │ └─DcmToNiiLog.txt
│ ├─BIDSConvert.log
│ └─FileTree.txt
├─sub-Test002/
│ └─ses-001/
│ ├─dix/
│ │ ├─sub-Test002_ses-001_stk-DIXON1_megre_imag.json
│ │ ├─sub-Test002_ses-001_stk-DIXON1_megre_imag.nii.gz
│ │ ├─sub-Test002_ses-001_stk-DIXON1_megre.json
│ │ ├─..<<34>>..
│ │ ├─sub-Test002_ses-001_stk-DIXON5_megre_ph.nii.gz
│ │ ├─sub-Test002_ses-001_stk-DIXON5_megre_real.json
│ │ └─sub-Test002_ses-001_stk-DIXON5_megre_real.nii.gz
│ ├─dwi/
│ │ ├─sub-Test002_ses-001_stk-DTI1_dwi_dti.bval
│ │ ├─sub-Test002_ses-001_stk-DTI1_dwi_dti.bvec
│ │ ├─sub-Test002_ses-001_stk-DTI1_dwi_dti.json
│ │ ├─..<<14>>..
│ │ ├─sub-Test002_ses-001_stk-DTI5_dwi_dti.bvec
│ │ ├─sub-Test002_ses-001_stk-DTI5_dwi_dti.json
│ │ └─sub-Test002_ses-001_stk-DTI5_dwi_dti.nii.gz
│ ├─raw/
│ │ ├─101_20221213173147_20221212-set2_Ph_20221212-set2_SURVEY_2_LEG-1_GR.json
│ │ ├─101_20221213173147_20221212-set2_Ph_20221212-set2_SURVEY_2_LEG-1_GR.nii.gz
│ │ ├─201_20221213173147_20221212-set2_Ph_20221212-set2_SURVEY_2_LEG-2_GR.json
│ │ ├─..<<432>>..
│ │ ├─805_20221213173147_20221212-set2_Ph_20221212-set2_WIP_DTI5_SE.json
│ │ ├─805_20221213173147_20221212-set2_Ph_20221212-set2_WIP_DTI5_SE.nii.gz
│ │ └─DcmToNiiLog.txt
│ ├─BIDSConvert.log
│ └─FileTree.txt
├─sub-viditest/
│ └─ses-001/
│ ├─dix/
│ │ ├─sub-viditest_ses-001_stk-DIXON1_megre_dix_imag.json
│ │ ├─sub-viditest_ses-001_stk-DIXON1_megre_dix_imag.nii.gz
│ │ ├─sub-viditest_ses-001_stk-DIXON1_megre_dix.json
│ │ ├─..<<42>>..
│ │ ├─sub-viditest_ses-001_stk-DIXON6_megre_dix_ph.nii.gz
│ │ ├─sub-viditest_ses-001_stk-DIXON6_megre_dix_real.json
│ │ └─sub-viditest_ses-001_stk-DIXON6_megre_dix_real.nii.gz
│ ├─dwi/
│ │ ├─sub-viditest_ses-001_stk-DTI1_dwi_dti.bval
│ │ ├─sub-viditest_ses-001_stk-DTI1_dwi_dti.bvec
│ │ ├─sub-viditest_ses-001_stk-DTI1_dwi_dti.json
│ │ ├─..<<18>>..
│ │ ├─sub-viditest_ses-001_stk-DTI6_dwi_dti.bvec
│ │ ├─sub-viditest_ses-001_stk-DTI6_dwi_dti.json
│ │ └─sub-viditest_ses-001_stk-DTI6_dwi_dti.nii.gz
│ ├─quant/
│ │ ├─sub-viditest_ses-001_stk-T21_mese_t2.json
│ │ ├─sub-viditest_ses-001_stk-T21_mese_t2.nii.gz
│ │ ├─sub-viditest_ses-001_stk-T22_mese_t2.json
│ │ ├─..<<6>>..
│ │ ├─sub-viditest_ses-001_stk-T25_mese_t2.nii.gz
│ │ ├─sub-viditest_ses-001_stk-T26_mese_t2.json
│ │ └─sub-viditest_ses-001_stk-T26_mese_t2.nii.gz
│ ├─raw/
│ │ ├─101_20230103162213_20230103-viditest_Ph_20230103-viditest_SUR-PLAN_GR.json
│ │ ├─101_20230103162213_20230103-viditest_Ph_20230103-viditest_SUR-PLAN_GR.nii.gz
│ │ ├─301_20230103162213_20230103-viditest_Ph_20230103-viditest_SUR-COR-1_GR.json
│ │ ├─..<<697>>..
│ │ ├─806_20230103162213_20230103-viditest_Ph_20230103-viditest_T2-6_RM_e9.json
│ │ ├─806_20230103162213_20230103-viditest_Ph_20230103-viditest_T2-6_RM_e9.nii.gz
│ │ └─DcmToNiiLog.txt
│ ├─FileTree.txt
│ └─sub-viditest_ses-001_BIDSConvert.log
├─DcmToNii_030123-191446.log
├─DcmToNii_040123-085253.log
├─DcmToNii_050123-170902.log
├─DcmToNii_271222-131841.log
├─DcmToNii_281222-091810.log
└─FileTree.txt