Full leg AI muscle segmentation using CNN
Automated muscle segmentation
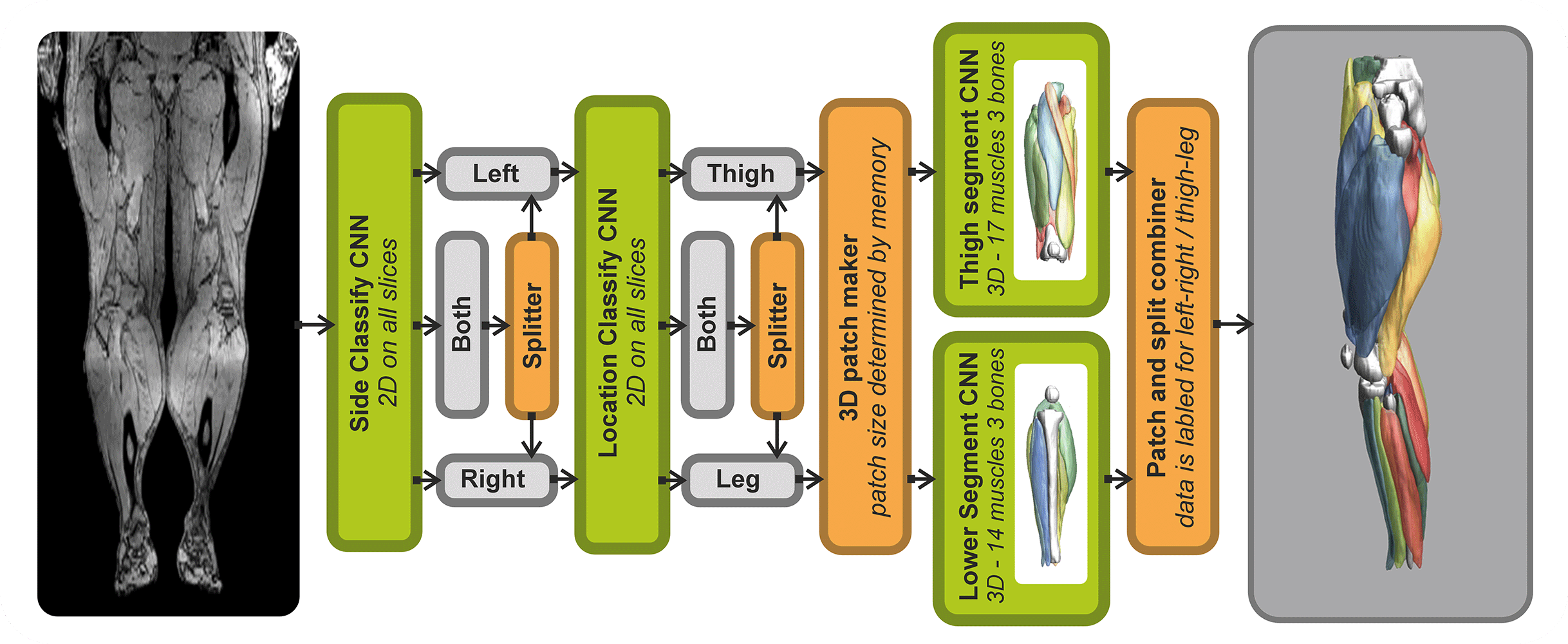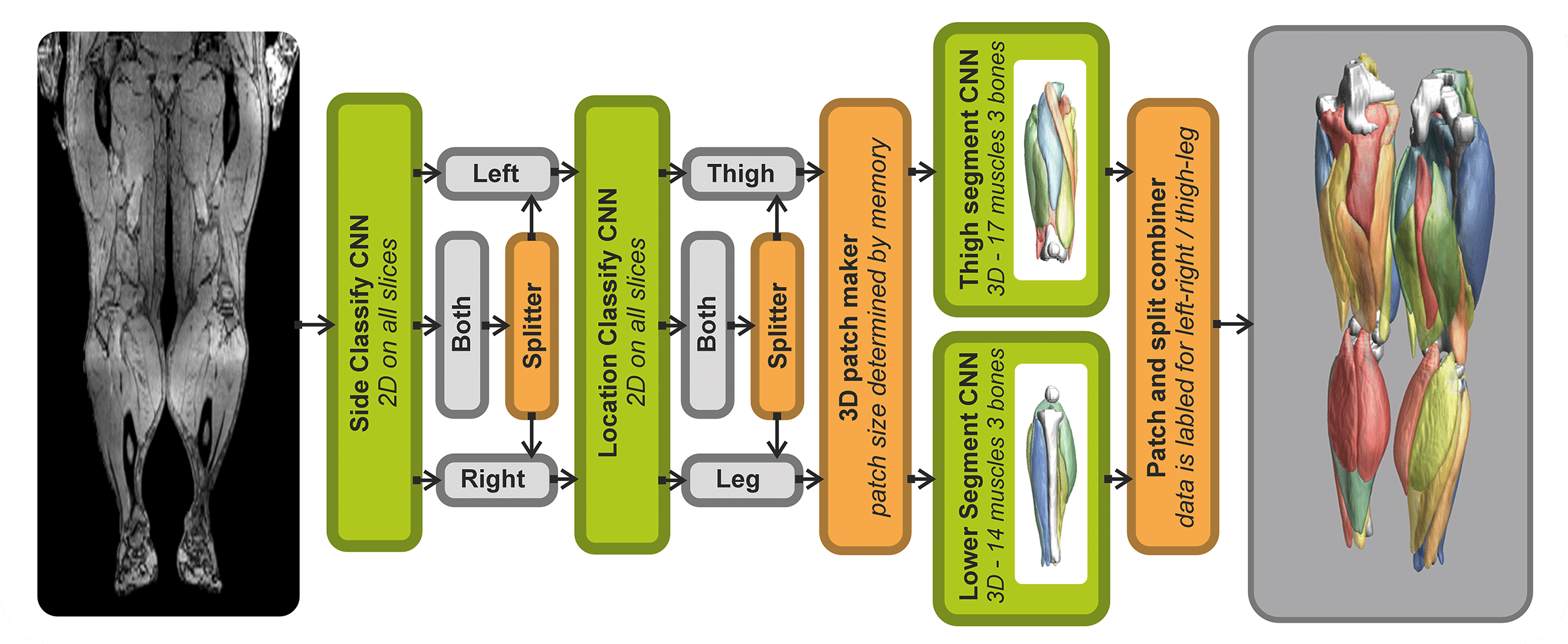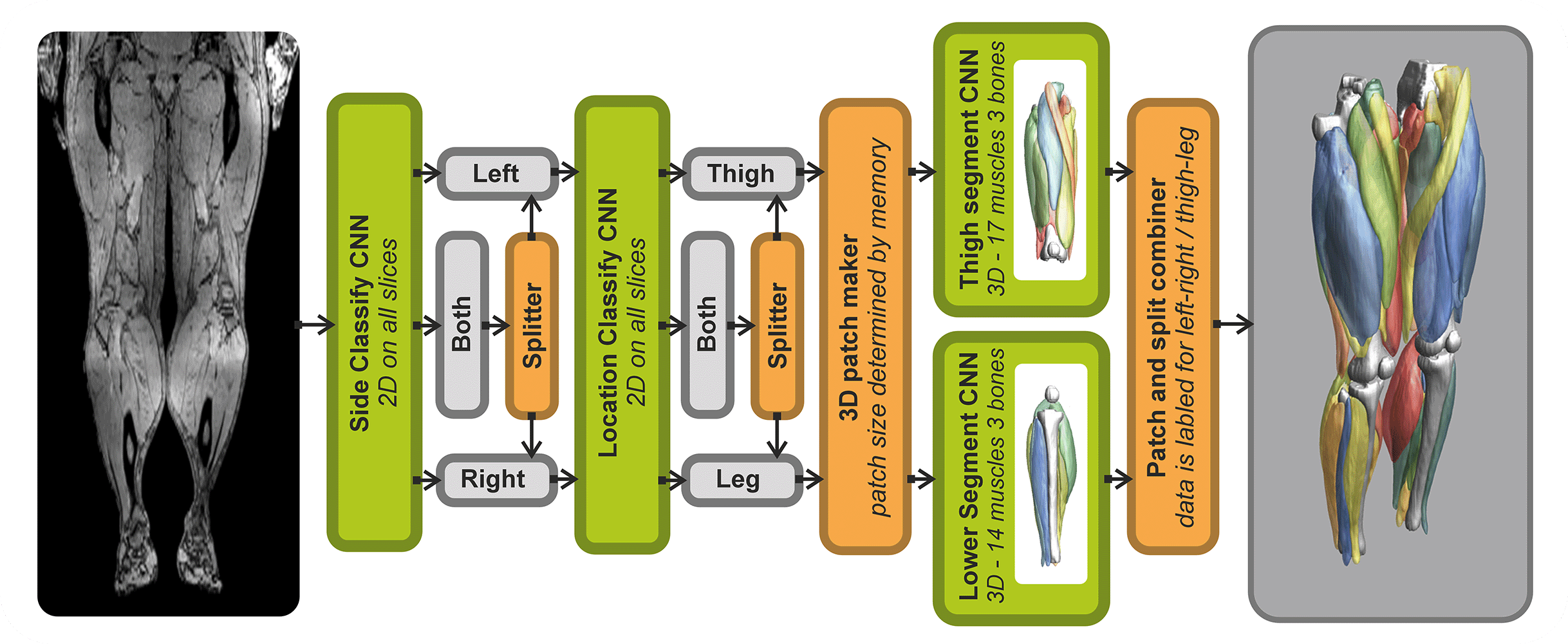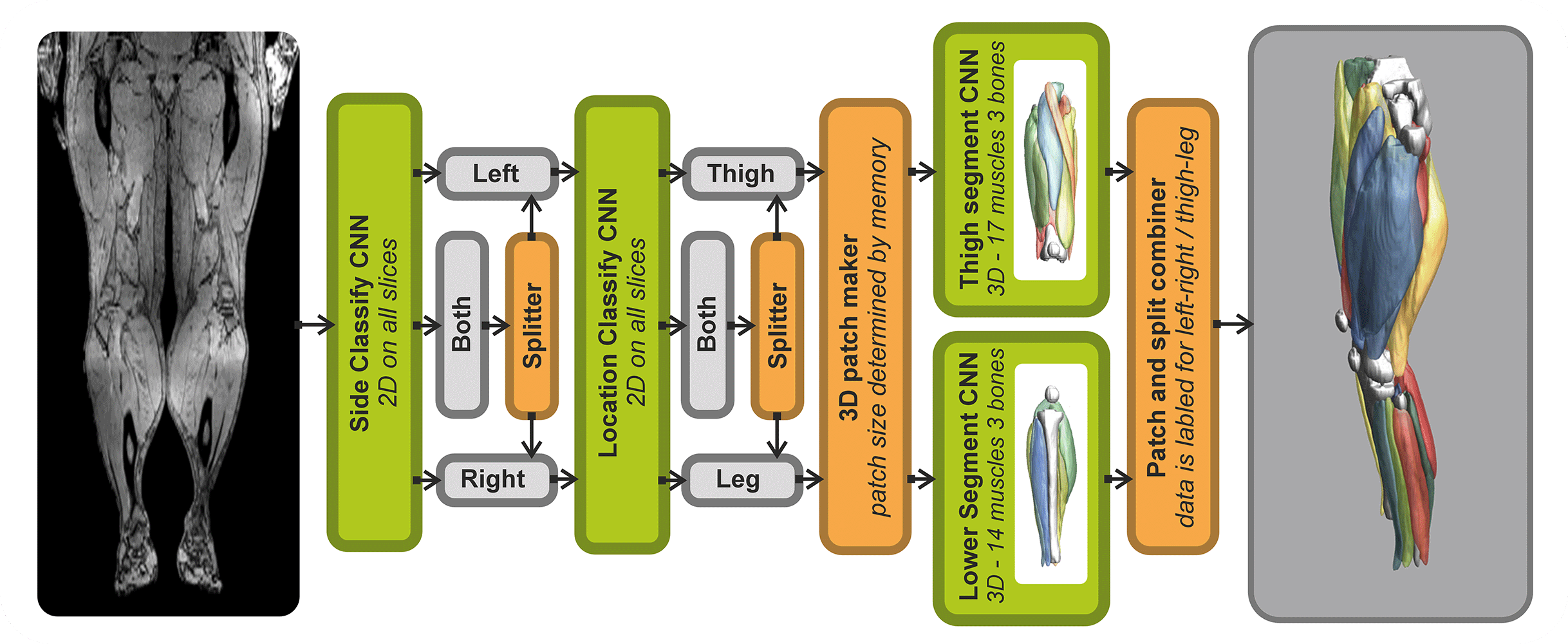
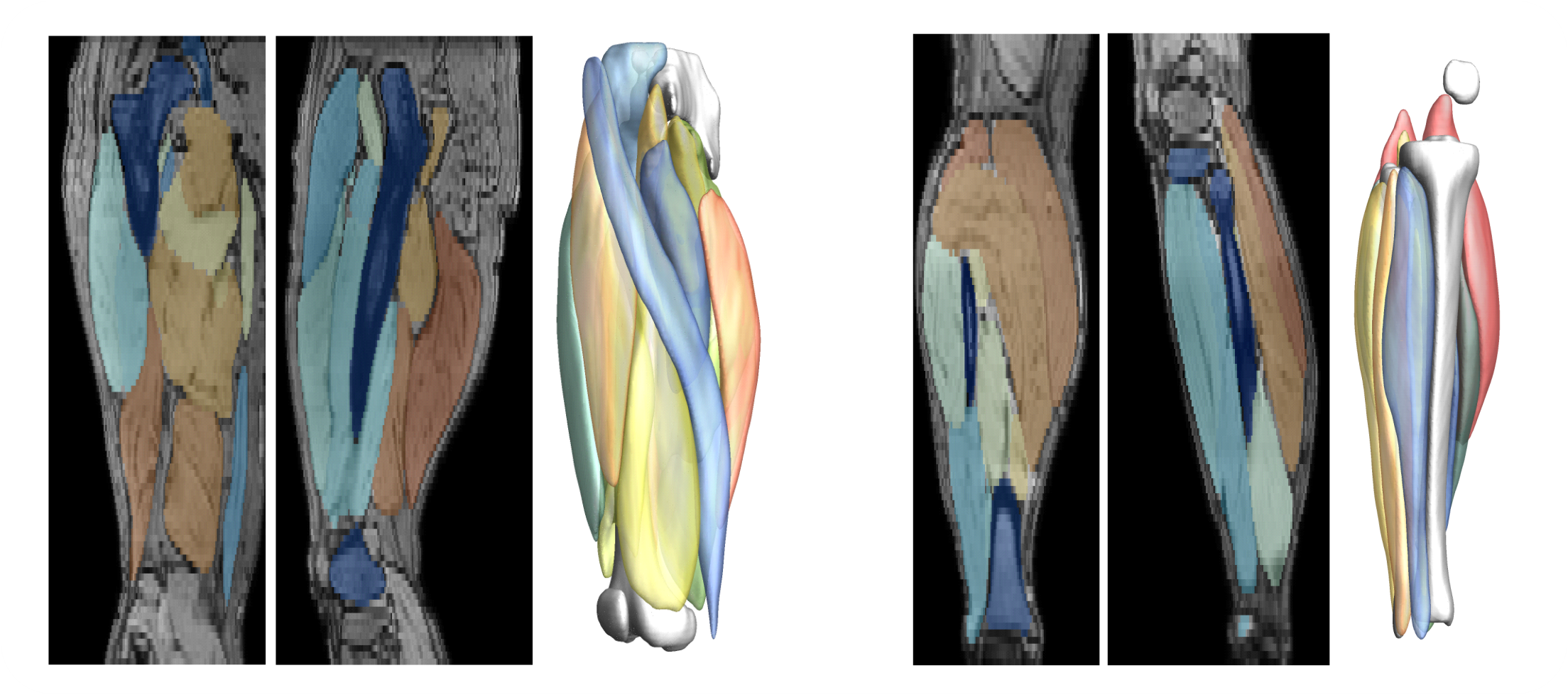
The SegmentData function within the QMRITools package is designed to automatically segment lower extremity muscles of healthy subjects of the hip, thigh, and leg. This function is an essential tool in the Segmentationtools package, leveraging advanced neural networks for precise muscle segmentation.
Function Overview for muscle segmentation
SegmentData employs four neural networks:
- Side and Location Recognition: Two networks identify the side (left/right) and location (hip/thigh/leg) of the muscles.
- Muscle Segmentation: Two networks perform the detailed segmentation of the muscles.
The networks are trained on 20 manually segmented thigh and leg datasets and currently operate on out-phase gradient echo images, which can be acquired or reconstructed using Dixon-based methods. The implementation supports any data size, with optimal performance on images with an in-plane resolution of 1-2.5 mm and a slice thickness of 5-10 mm. The latest trained networks are available as .wlnet files within the QMRItools package package, and .ONNX files can be requested.
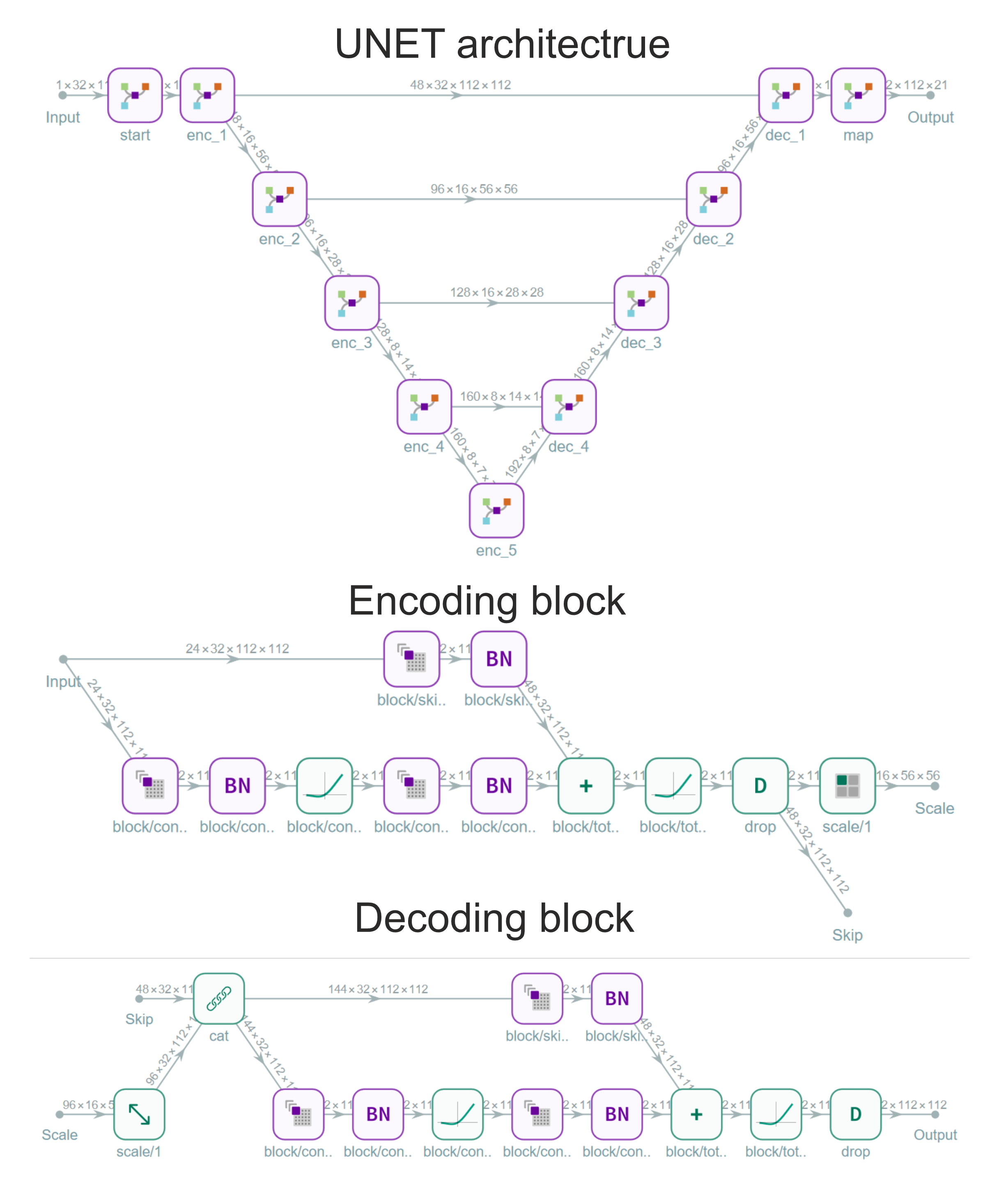
Architecture and Training
The segmentation networks are based on the UNET architecture with ResNet convolution blocks that contain short skip connections. The block diagrams of the encoding and decoding blocks used in our UNET architecture are shown below.
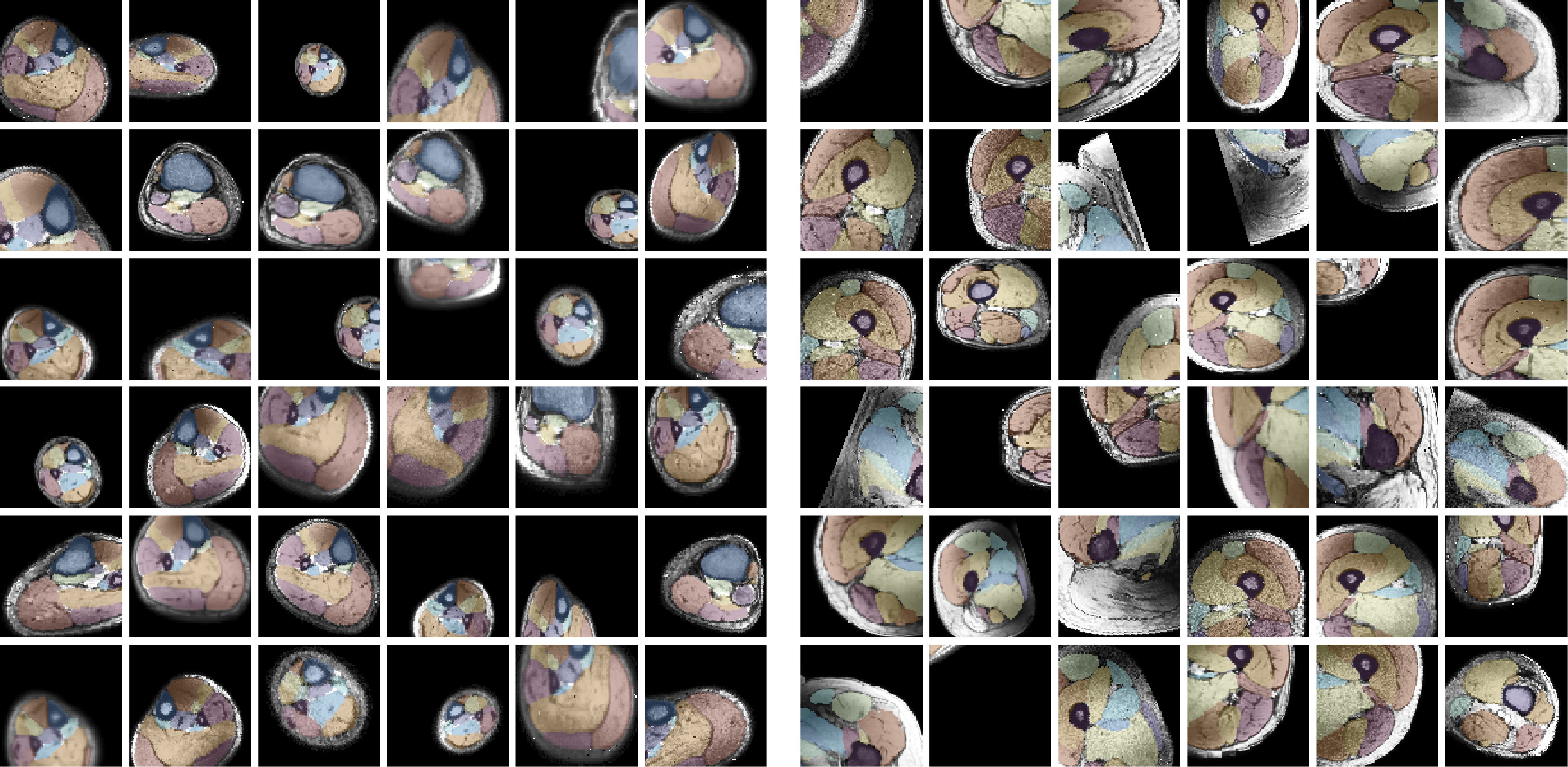
Data Augmentation and Training
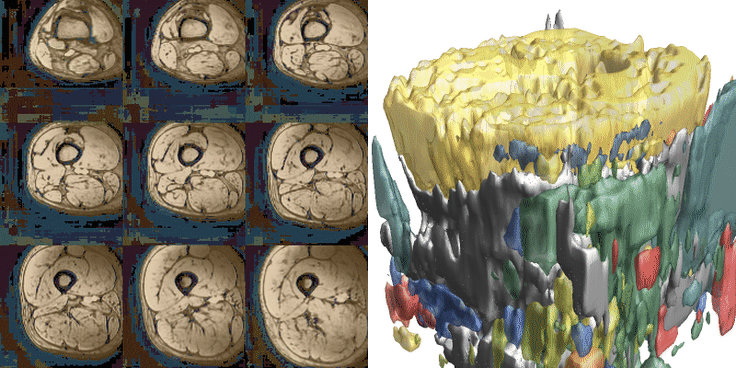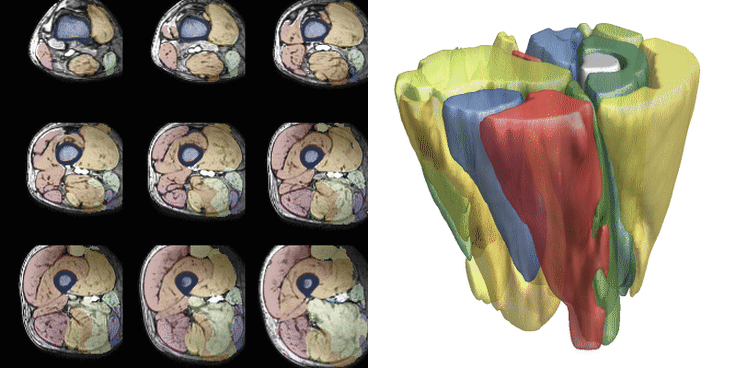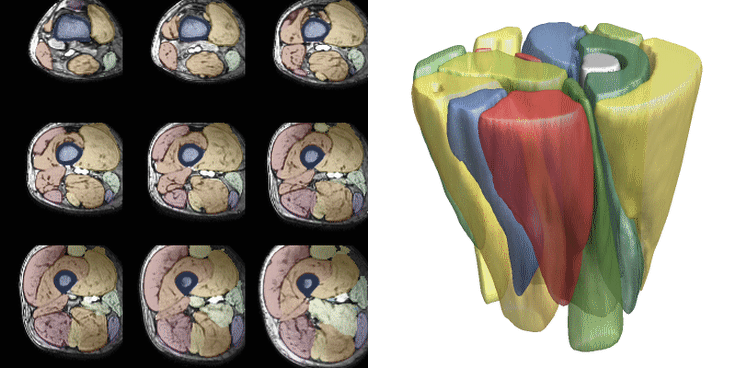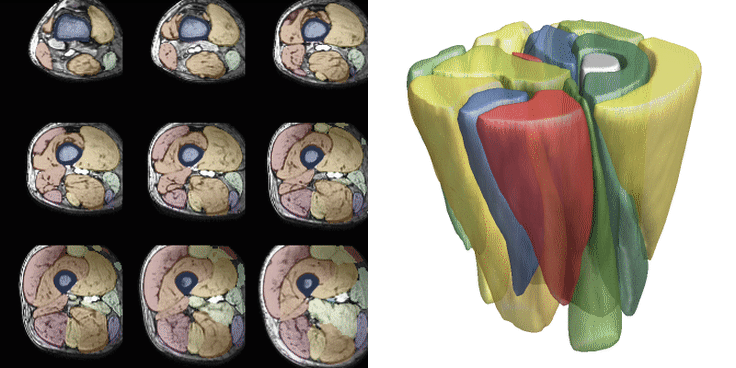
For training, data is heavily augmented using various techniques such as scale, skew, rotation, translation, noise, sharpen, contrast, and brightness adjustments. Training is conducted with a batch size of 2 and a patch size of 40x96x96 voxels, with 512 datasets seen per epoch. Patches are selected after data augmentation. Training typically continues for 200-300 epochs, taking approximately 8-12 hours. Below are examples of 36 different patches with augmentation selected from the same dataset. The resulting segmentations from both networks are displayed. The training for the upper leg is shown below.
Using SegmentData as a Script
No Mathematica licence? Not a problem it also works without. How this works you can read here.
To use SegmentData as a script, you can refer to the README available on GitHub. The README provides detailed instructions on how to implement the function, ensuring users can effectively utilize the tool for their specific research needs.
Features Include
- Automated region recognition
- Memory optimized segmentation
- CPU and GPU support
- Automated patching and merging
- Standardized muscle labels