How to use Muscle-BIDS in QMRITools
QMRITools and Muscle-BIDS
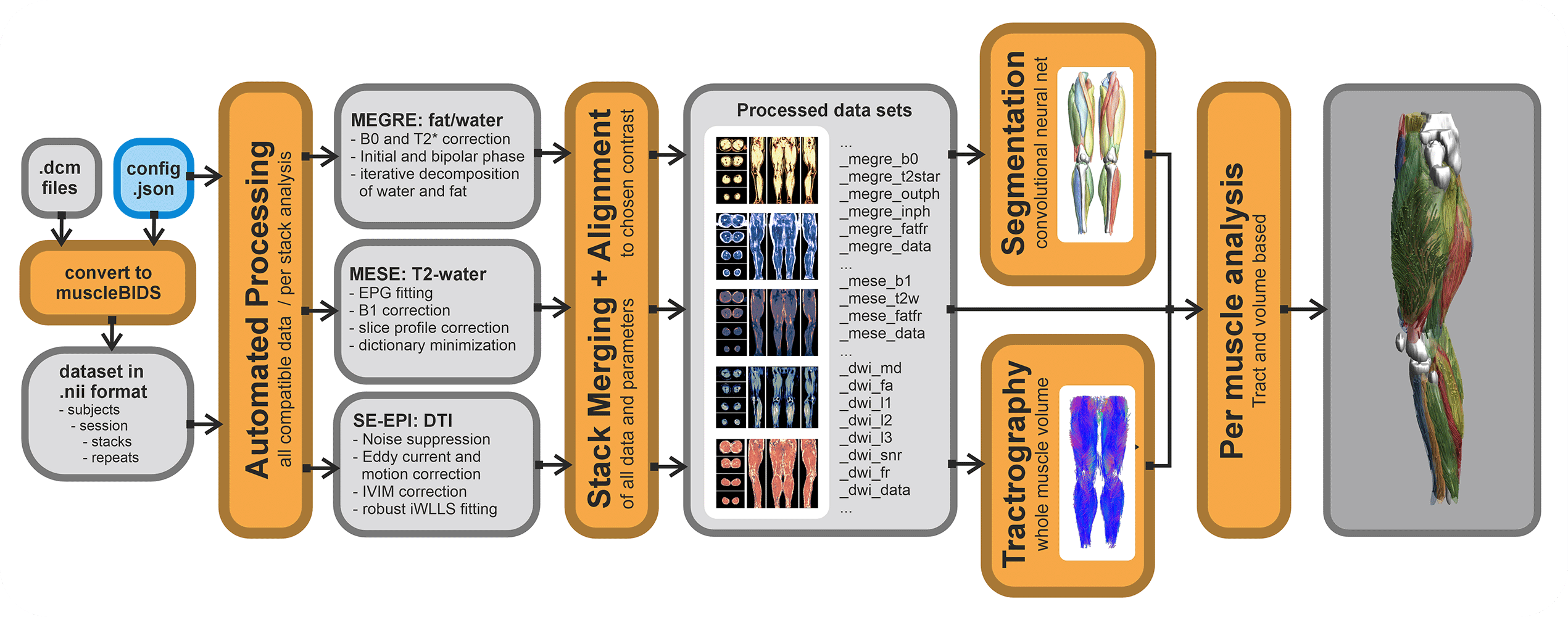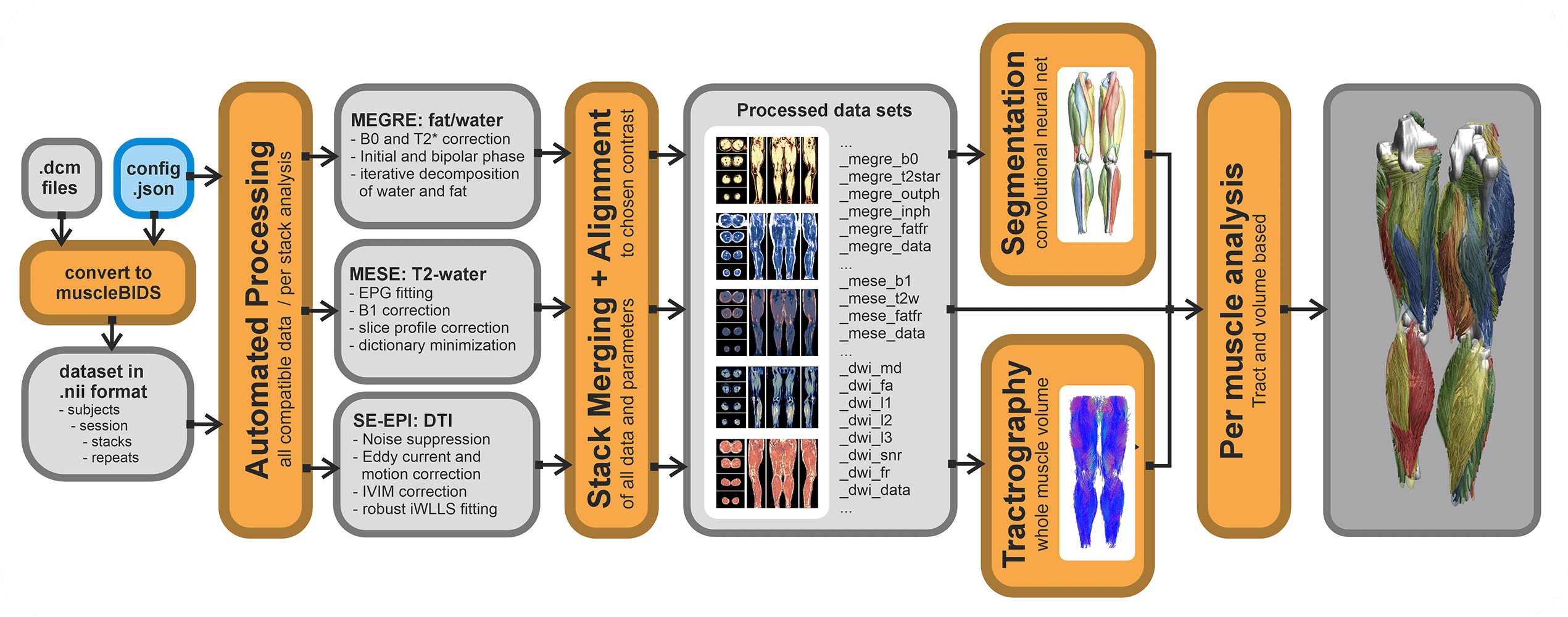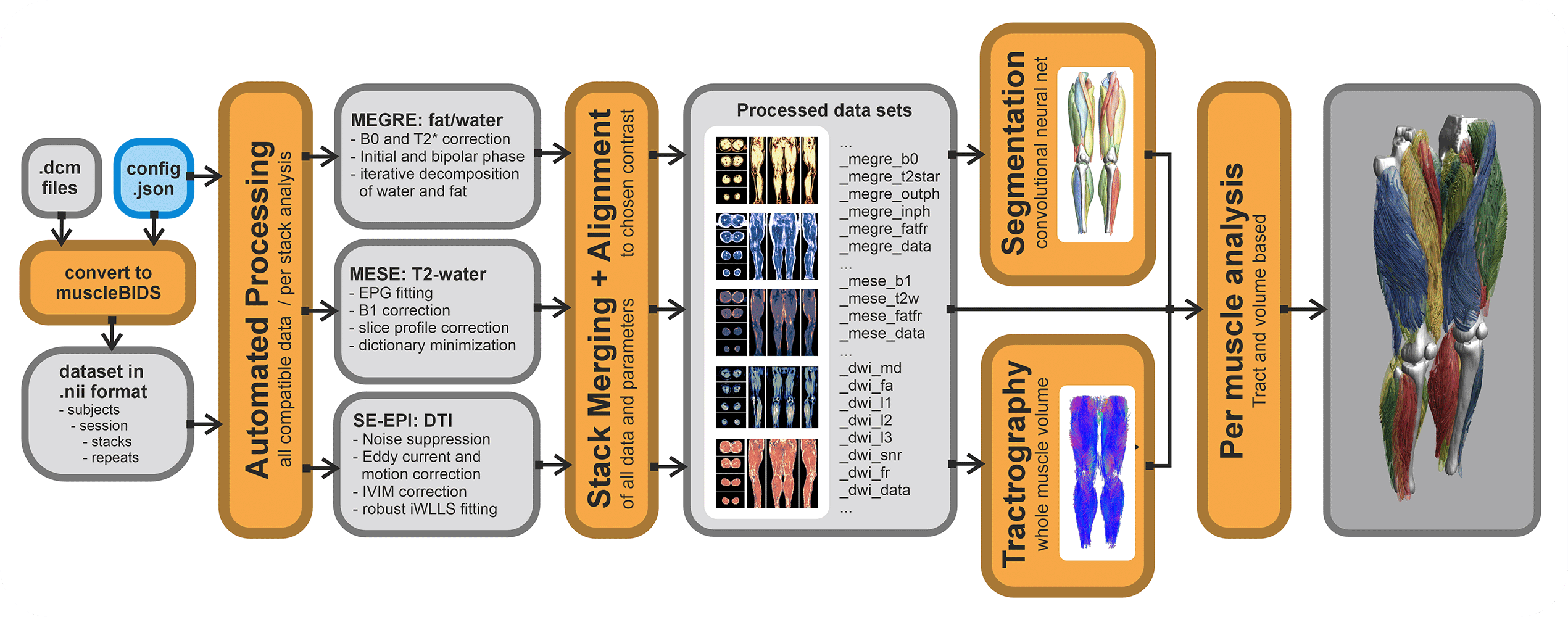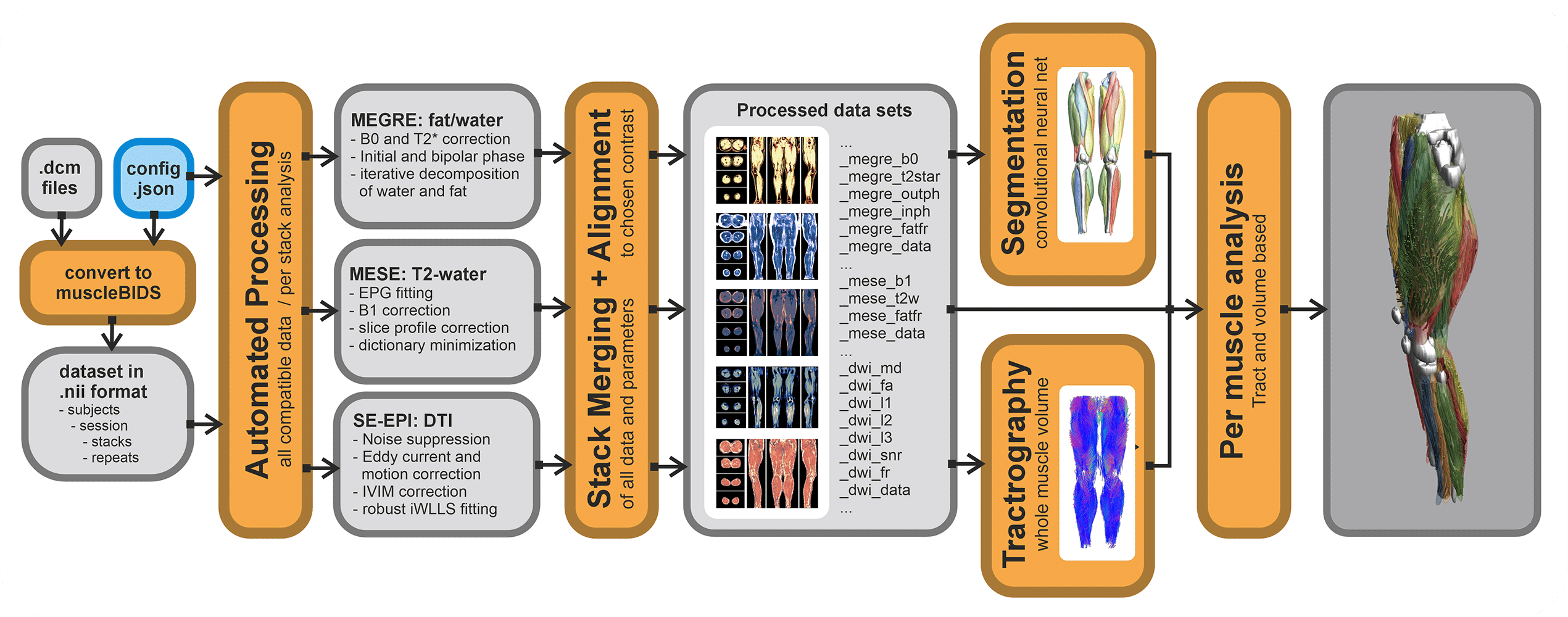
Automated processing only works if all input data is well-curated and defined. The general rule of garbage in is garbage out applies here as well. QMRITools allows for the automatization of most processing steps IF the data is standardized and well-described.
Currently, a config file for a dataset needs to be defined. This stores the data to be expected and in which form. Based on the data described the correct (pre-)processing scripts will be selected and performed. If expected data is not found the processing step will be skipped. Also, if the data is not described in the config it will be ignored. The config file can be general for the entire study folder, or can be defined on a per subject level.
Muscle-BIDS config
The configuration file can be present on each level of the Muscle-BIDS dataset. The best way is to define one config file in the root of the dataset. However, when a config file is also placed on the subject of session level this is used ignoring the global config. This allows making subject- or session-specific exceptions. An example of a config file for a dataset that contains Dixon, DWI and T2 data acquired over multiple stacks is given here».
Conversion of DICOM to raw Muscle-BIDS NIfTI
Putting data in a Muscle_BIDS data structure can be done manually, however, this can be very time-consuming. Therefore, QMRITools has some automated scripts that can do this. Conversion from Dicom to nifty can be done in many different ways and many tools exist. It is not needed to use the QMRITools functions for this. Currently, it is only tested for very specific acquisition protocols and custom studies custom scripts likely have to be written. Based on the existing scripts this should not be too difficult.
For Dicom to Nifti conversion, QMRITools uses dcm2niix which already converts most data to BIDS-compliant nifty (function: BidsDcmToNii). These conversions are stored in the source data folder "raw". Next, a second script collects the needed data and converts, moves and renames it to Muscle-BIDS-compliant files and folders (function: MuscleBidsConvert). This is done based on the user-defined config file. Both function loop over all folders and ignore the ones already processed.
Processing of Muscle-BIDS data
If the dataset is set up properly and the config file is defined data processing of known data types is done fully automated (function: MuscleBidsProcess). The processing script will go through all folders finds the needed data and process all data for which the needed input is present. Data already processed will be skipped. Reprocessing of data can be forced for all data, data processed before a specific data or data processing with an old QMRITools version. To keep track of all this QMRITools creates additional "*.json" files with the suffix "_check". If these files are not present data will be reprocessed. After data processing extra information can be obtained from the generated data such as muscle segmenation based on the "_megre_dix_outph" data (function: MuscleBidsSegment) or whole volume fiber tractography using the "_dwi_dti_tens" data (function: MuscleBidsTractography).
Currently, the following automated processing scripts exist:
- Dixon reconstruction using “_megre” data.
- IVIM-DTI preprocessing and modelling using “_dwi” data.
- Water T2 calculation using “_mese” data using EPG modelling.
- Merging multiple stacks into one dataset.
- Whole volume muscle fiber tractography.
- Automated leg and thigh muscle segmentation
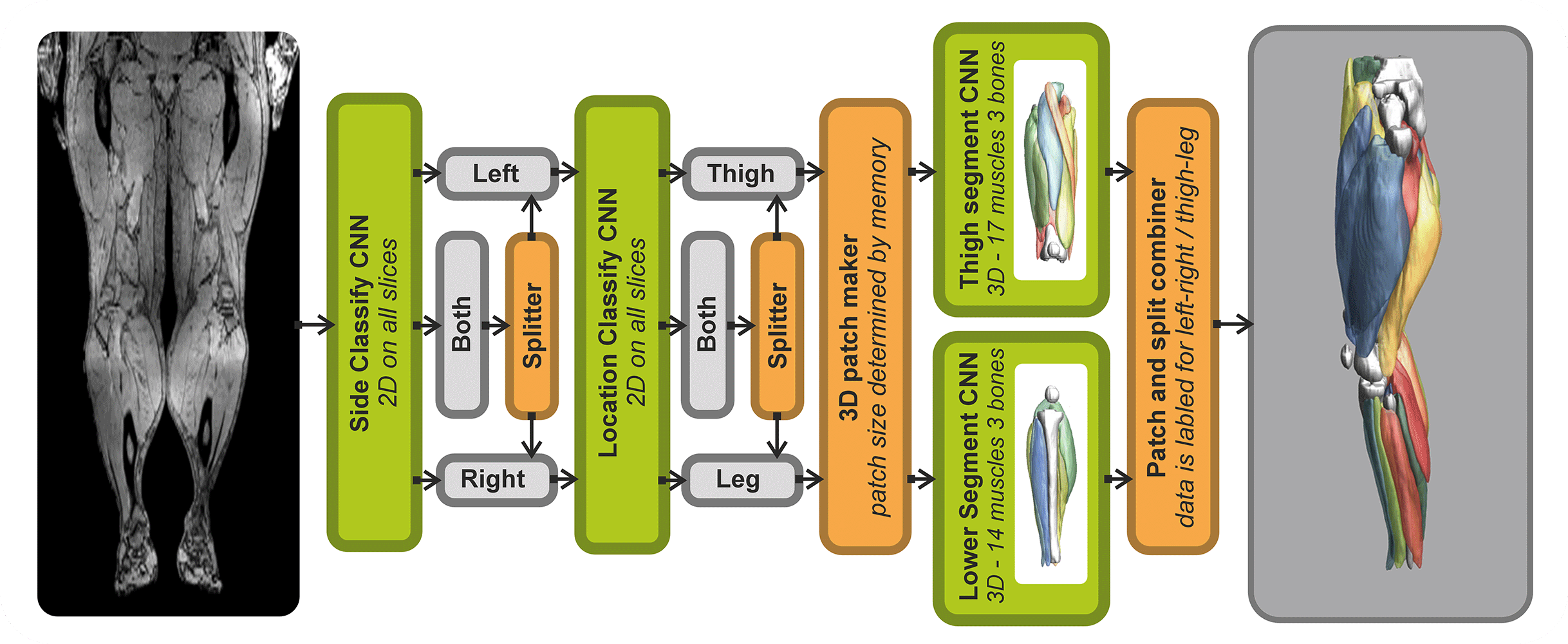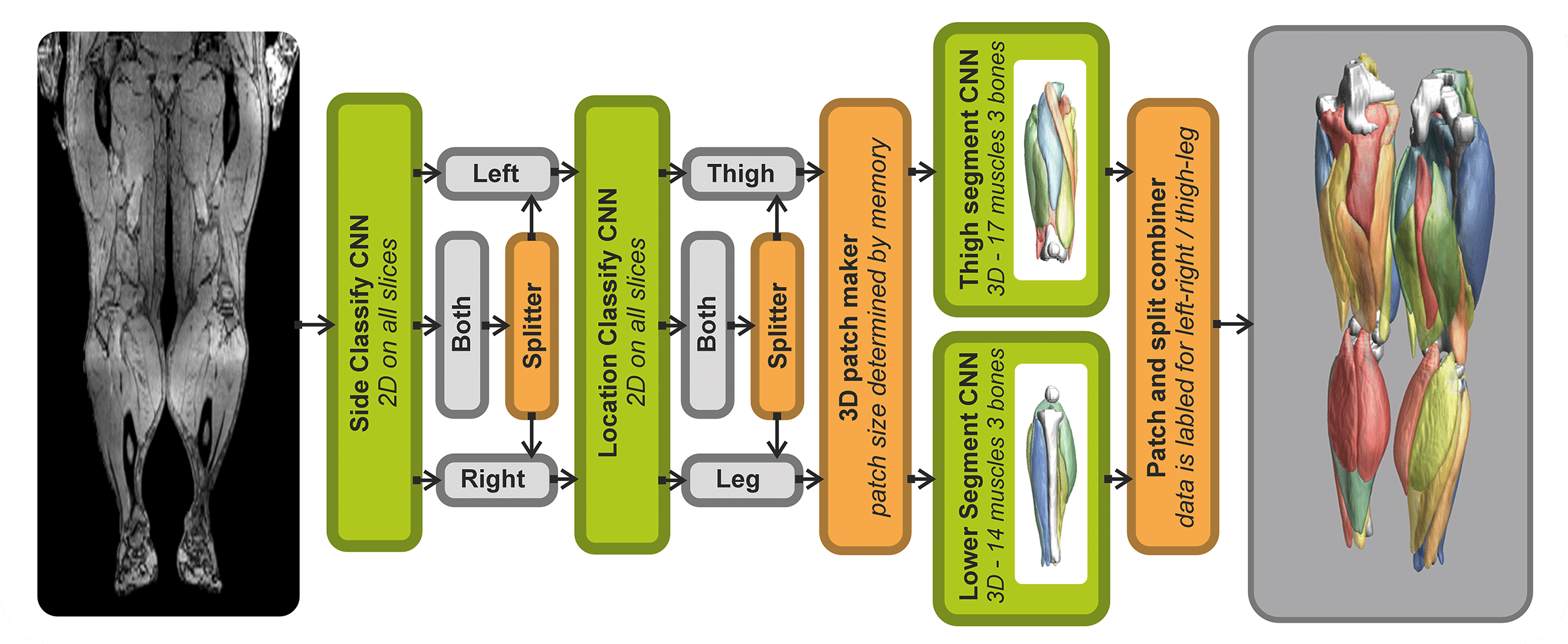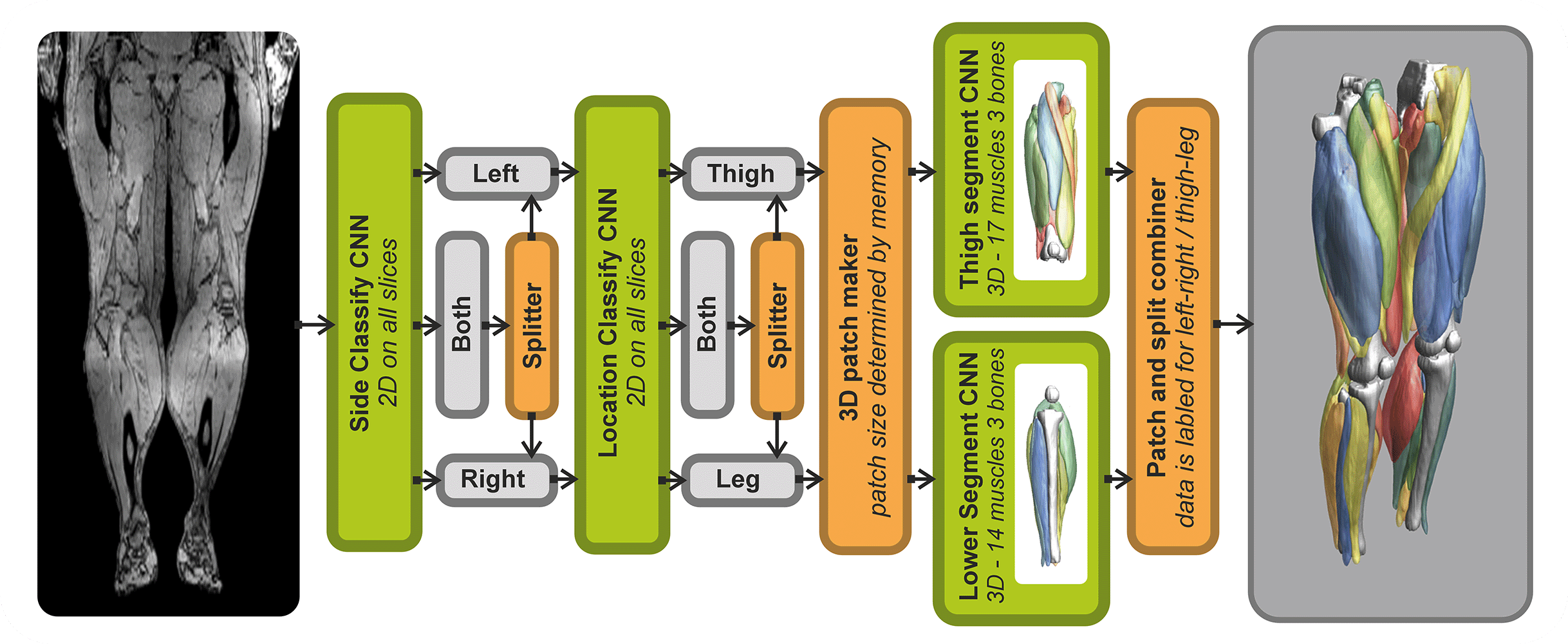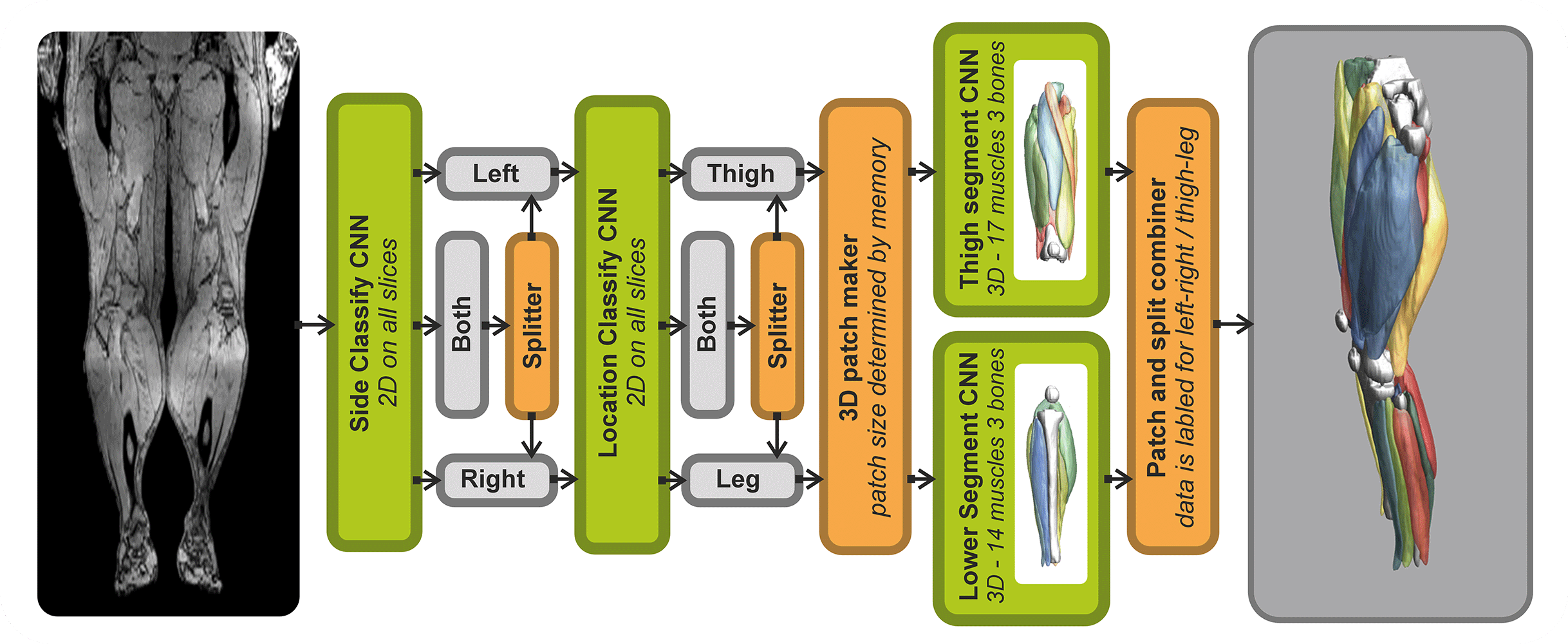
Analysis of Muscle-BIDS data
Based on the Muscle-BIDS outputs the generated qMRI parameters can be analyzed of used for muscle evaluation. Once all muscle segmentation and tracts are known per muscle qMRI parameters can be summarized. The following “general” scripts that can summarize qMRI analysis for statistical analysis are being developed:
- Masked-based qMRI parameter extraction to xls.
- Tract-based qMRI parameter extraction to xls.
- Muscle architecture description to xls.